Advanced Data Managment
National Data Management Center for Health (NDMC) at EPHI
Data Manipulation and Claning using dplyr() package
What is Tidyverse?
- The
tidyverseis a collection of R packages designed for data science.- All packages share an underlying design philosophy, grammar, and data structures.
- All packages included in
tidyverseare automatically installed when installing thetidyversepackage:
- Then to work functions under tidyverse package we must always load the package into the workplace.
- Some packages under tidyverse are considered
core packagesand others calledfriend packages.
install.packages(“tidyverse”)
Core tidyverse
-
tibble, for tibbles, a modern re-imagining of data frames -
readr, for data import -
tidyr, for data tidying -
ggplot2, for data visualization -
dplyr, for data manipulation -
stringr, for strings -
forcats, for factors -
purrr, for functional programming
Friends for data import or export (beyond readr)
-
readxl, for xls and xlsx files -
haven, for SPSS, SAS, and Stata files -
jsonlite, for JSON -
xml2, for XML -
httr, for web APIs -
rvest, for web scraping -
DBI, for databases
Friends for date wrangling
-
lubridateandhms, for date/times
Friends for modeling
-
modelrandbroomfor model/tidy data
For this training we manly use the following Core Packages

Intro to dplyr package
dplyris part oftidyverseprovides a grammar (the verbs) for data manipulation.-
The key operator and the essential verbs are:
Function Description Operates on filter()pick rows matching criteria rows slice()pick rows using indices rows arrange()reorder rows rows select()pick columns by name columns mutate()add new variables columns summarise()reduce variables to values groups of rows relocate()to change column positions columns … many more.
%>%or|>: the “pipe” operator used to connect multiple verb actions together into a pipeline.
Tools → Global Options → Code → Editing → Use Native Pipe Operator (|>)
Select Columns from a Dataset select():
select(): To extract variables

select()∼ columnsselect columns (variables)
no quotes needed around variable names
can be used to rearrange columns
uses special syntax that is flexible and has many options
Note that the column names are not quoted; you access the column name as if you are calling the name of an object or variable
Ways to Use select() in dplyr
| Method | Description | Example | |
|---|---|---|---|
| using column name | Select specific columns by their names. | select(col1, col2) |
|
| By position | Select columns by their positions. | select(1, 3) |
|
| Using a range | Use : to select columns |
select(col1:col5); select(2:4) |
|
| Exclude columns | Use - to exclude specific columns |
select(-col3); select(-(2:4));``select(!starts_with("A")) |
|
| Use pattern | Use helper functions based on patterns. |
select(starts_with("prefix"))select(ends_with("suffix"))select(contains("text"))
|
|
| Select by type | Use where() to select based on type or condition. |
select(where(is.numeric))select(where(~ n_distinct(.) > 5))
|
|
Select columns using matches()
|
Use a regular expression to select columns matching a specific pattern. | select(matches("col[1-3]")); ``select(matches("^var\\d+")) |
|
| Select all columns except some | Use everything() to re-order or select all columns except specific ones. |
select(col1, everything())select(-starts_with("temp"))
|
|
| Rearrange columns | Move specific columns to the front while retaining all others. | select(col1, col3, everything()) |
About the data
Data from the CDC’s Youth Risk Behavior Surveillance System (YRBSS)
- complex survey data
- national school-based survey conducted by CDC
- monitors six categories of health-related behaviors
- that contribute to the leading causes of death and disability among youth and adults
- including alcohol & drug use, unhealthy & dangerous behaviors, sexuality, and physical activity
- the data in
yrbss_demo.csvare a subset of data in the R packageyrbss
- We can have a look at the data and its structure by using the
glimpse()function from thedplyrpackage.
Pipe perator (|>)
Pipes in R look like
|>and strings together commands to be performed sequentiallyThe pipe passes the data frame output that results from the function right before the pipe to input it as the first argument of the function right after the pipe.
This nesting is not a natural way to think about a sequence of operations.
The
|>operator allows you to string operations in a left-to-right fashion.
Advantages of Pipe oprator
- Pipes used to reduce multiple steps, that can be hard to keep track of.
- less redundant code
- Easy to read and write because functions are executed in order
- Difficult to read if too many functions are nested
- Look at the three syntax
select a column by name: select(col1, col2, col3, ...)
# A tibble: 20,000 × 3
age sex grade
<chr> <chr> <chr>
1 15 years old Female 10th
2 17 years old Female 12th
3 18 years old or older Male 11th
4 15 years old Male 10th
5 14 years old Male 9th
6 17 years old Male 9th
7 16 years old Male 11th
8 17 years old Male 12th
9 18 years old or older Male 12th
10 14 years old Male 10th
# ℹ 19,990 more rowsSelecting column ranges with :
- The
:operator selects a range of consecutive variables:
# A tibble: 3 × 4
age sex grade race4
<chr> <chr> <chr> <chr>
1 15 years old Female 10th White
2 17 years old Female 12th White
3 18 years old or older Male 11th Hispanic/Latino- We can also specify a range with column numbers:
Excluding columns with ! or -
The exclamation point negates a selection:
# A tibble: 2 × 7
age sex grade race4 race7 bmi stweight
<chr> <chr> <chr> <chr> <chr> <dbl> <dbl>
1 15 years old Female 10th White White 17.2 54.4
2 17 years old Female 12th White White 20.2 57.2To drop a range of consecutive columns, we use, for example,!age:grade:
# A tibble: 2 × 5
record race4 race7 bmi stweight
<dbl> <chr> <chr> <dbl> <dbl>
1 931897 White White 17.2 54.4
2 333862 White White 20.2 57.2To drop several non-consecutive columns, place them inside !c():
Helper functions: starts_with(), ends_with() and contains()
- These two helpers work exactly as their names suggest!
starts_with()
# A tibble: 2 × 3
record race4 race7
<dbl> <chr> <chr>
1 931897 White White
2 333862 White Whiteends_with()
contains()
-
contains()helps select columns that contain a certain string:
# A tibble: 6 × 5
sex record grade race4 race7
<chr> <dbl> <chr> <chr> <chr>
1 Female 931897 10th White White
2 Female 333862 12th White White
3 Male 36253 11th Hispanic/Latino Hispanic/Latino
4 Male 1095530 10th Black or African American Black or African American
5 Male 1303997 9th All other races Multiple - Non-Hispanic
6 Male 261619 9th All other races <NA> Another helper function, everything()
- matches all variables that have not yet been selected.
# A tibble: 3 × 8
bmi record age sex grade race4 race7 stweight
<dbl> <dbl> <chr> <chr> <chr> <chr> <chr> <dbl>
1 17.2 931897 15 years old Female 10th White White 54.4
2 20.2 333862 17 years old Female 12th White White 57.2
3 NA 36253 18 years old or older Male 11th Hispanic/Latino Hisp… NA It is often useful for establishing the order of columns.
- But this would be painful for larger data frames, data frame. In such a case, we can use
everything().
- This helper can be combined with many others.
Code
# A tibble: 3 × 8
record race4 race7 age sex grade bmi stweight
<dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl>
1 931897 White White 15 years old Fema… 10th 17.2 54.4
2 333862 White White 17 years old Fema… 12th 20.2 57.2
3 36253 Hispanic/Latino Hispanic/Latino 18 years ol… Male 11th NA NA - You can also select columns based on their data type using
select_if(). - The common data types to be called are:
is.character,is.double,is.factor,is.integer,is.logical,is.numeric.
Code
Rows: 20,000
Columns: 3
$ record <dbl> 931897, 333862, 36253, 1095530, 1303997, 261619, 926649, 1309…
$ bmi <dbl> 17.1790, 20.2487, NA, 27.9935, 24.4922, NA, 20.5435, 19.2555,…
$ stweight <dbl> 54.43, 57.15, NA, 85.73, 66.68, NA, 70.31, 58.97, 123.38, NA,…Summary for Select() function
- There are five ways to select variables in
select(data, ...):
-
By position:
yrb_data |> select(1, 2, 4)oryrb_data |> select(1:2). -
By name:
yrb_data |> select(age, sex), oryrb_data |> select(age:race4). -
By function of name:
yrb_data |> select(starts_with("r")), oryrb_data |> select(ends_with("e")). -
By type:
yrb_data |> select(where(is.numeric)),oryrb_data |> select(where(is.character)). -
By any combination of the above using the Boolean operators !, &, and |:
-
yrb_data |> select(!where(is.numeric)):selects all non-factor variables. -
yrb_data |> select(where(is.numeric) & contains("i")):selects all numeric variables that contains ‘i’.
-
-
Also the following are other important function
-
_all()if you want to apply the function to all columns -
_at()if you want to apply the function to specific columns (specify them withvars()) -
_if()if you want to apply the function to columns of a certain characteristic (e.g. data type) -
_with()if you want to apply the function to columns and include another function within it
-
Rows: 20,000
Columns: 8
$ record <dbl> 931897, 333862, 36253, 1095530, 1303997, 261619, 926649, 1309…
$ age <chr> "15 years old", "17 years old", "18 years old or older", "15 …
$ sex <chr> "Female", "Female", "Male", "Male", "Male", "Male", "Male", "…
$ grade <chr> "10th", "12th", "11th", "10th", "9th", "9th", "11th", "12th",…
$ race4 <chr> "White", "White", "Hispanic/Latino", "Black or African Americ…
$ race7 <chr> "White", "White", "Hispanic/Latino", "Black or African Americ…
$ bmi <dbl> 17.1790, 20.2487, NA, 27.9935, 24.4922, NA, 20.5435, 19.2555,…
$ stweight <dbl> 54.43, 57.15, NA, 85.73, 66.68, NA, 70.31, 58.97, 123.38, NA,…- If we wanted only some of them renamed and kept, we could have used
select_at()which specifies columns withvars().
Code
Rows: 20,000
Columns: 3
$ SEX <chr> "Female", "Female", "Male", "Male", "Male", "Male", "Male", "Male"…
$ AGE <chr> "15 years old", "17 years old", "18 years old or older", "15 years…
$ BMI <dbl> 17.1790, 20.2487, NA, 27.9935, 24.4922, NA, 20.5435, 19.2555, 33.1…Filter cases from the dataset filter()
filter(): To extract cases
The function filter() is used to filter the dataset to return a subset of all rows that meet one or more specific conditions.
filter(dataframe, logical statement 1, logical statement 2, ...)
.png)

Ways to Use filter() in dplyr
| Method | Description | Example |
|---|---|---|
| By specific value | Filter rows where a column equals a specific value. |
filter(col1 == "value") filter(col1 != "value")
|
| By inequality | Filter rows based on inequality conditions. | filter(col1 > 10) |
| Using multiple conditions | Filter rows that satisfy multiple conditions. | filter(col1 > 10, col2 == "A") |
| With logical operators | Using AND (&) and using OR (|) |
|
| By range | Filter rows within a range of values using between(). |
filter(between(col1, 10, 20)) |
| By missing values | Filter rows with or without missing values. |
filter(is.na(col1))filter(!is.na(col1))
|
| By string pattern | Filter rows using string functions like grepl(). |
filter(grepl("pattern", col1)) |
Using case_when()
|
Apply conditional logic for complex filtering. | filter(case_when(col1 > 10 ~ TRUE, TRUE ~ FALSE)) |
Filtering based on exact character variable matches
- Note the use of the double equal sign
==rather than the single equal sign=.
# A tibble: 3 × 2
sex grade
<chr> <chr>
1 Male 9th
2 Male 9th
3 Male 9th # A tibble: 3 × 2
sex grade
<chr> <chr>
1 Male 11th
2 Male 10th
3 Male 9th - Similarly you can use the other operators:
-
filter(grade != "9th")will select everything except the grade 9 rows.
-
- If you want to select more than one category value you can use the
%in%operator.
- The
%in%operator used to deselect certain groups as well, using!%in%.
- To select all individuals with a bmi between 22 and 30, use:
Filtering based on multiple conditions
The filter option also allows AND and OR style filters:
filter(condition1, condition2)will return rows where both conditions are met.filter(condition1, !condition2)will return all rows where condition one is true but condition 2 is not.filter(condition1 | condition2)will return rows where condition 1 and/or condition 2 is met.filter(xor(condition1, condition2)will return all rows where only one of the conditions is met, and not when both conditions are met.
Code
# A tibble: 3 × 5
sex age bmi stweight grade
<chr> <chr> <dbl> <dbl> <chr>
1 Female 17 years old 20.2 57.2 12th
2 Male 15 years old 28.0 85.7 10th
3 Male 14 years old 24.5 66.7 9th Example with xor()
Filtering across multiple columns
| Method | Description | Example |
|---|---|---|
across() |
Filter rows based on conditions across multiple columns. | filter(across(starts_with("col"), ~ .x > 10)) |
filter_at() |
Filter rows based on selected columns using conditions. | filter_at(vars(col1, col2), all_vars(. > 10)) |
filter_if() |
Filter rows based on a condition for specific column types. | filter_if(is.numeric, all_vars(. > 10)) |
filter_all() |
Apply the same condition across all columns. | filter_all(all_vars(. > 0)) |
across() and any_vars()
|
Keep rows where any column satisfies a condition. | filter(across(starts_with("col"), any_vars(. > 10))) |
across() and all_vars()
|
Keep rows where all selected columns satisfy a condition. | filter(across(ends_with("score"), all_vars(. >= 50))) |
between() in multiple columns |
Filter rows where values fall within a range. | filter(across(c(col1, col2), ~ between(.x, 10, 20))) |
-
across()is the modern and flexible replacement forfilter_at(),filter_if(), andfilter_all(). -
all_vars()ensures all selected columns satisfy the condition. -
any_vars()ensures at least one selected column satisfies the condition.
filter_all()
-
Used to wrap the condition in any_vars().
- For example to find the string “symp” across all columns,
- The same can be done for numerical values: This code will retain any rows that has any value above 120:
-
any_vars()statement is equivalent to OR, and -
all_vars()is an equivalent for AND.
The below code will retain any rows where all values are below 15.
filter_if()
to find out all data rows where we NA in the first few columns use
filter_all(any_vars(is.na(.)))Using
filter_if()I can filter on character variables.
# A tibble: 6 × 8
record age sex grade race4 race7 bmi stweight
<dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl>
1 36253 18 years old or older Male 11th Hispanic/Latino Hisp… NA NA
2 261619 17 years old Male 9th All other races <NA> NA NA
3 180494 14 years old Male 10th Black or Africa… Blac… NA NA
4 31226 15 years old Male 10th All other races <NA> NA NA
5 109404 16 years old Male 11th White White NA NA
6 180968 15 years old Male 9th White White NA NA- Or
# A tibble: 6 × 8
record age sex grade race4 race7 bmi stweight
<dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl>
1 931897 15 years old Female 10th White White 17.2 54.4
2 333862 17 years old Female 12th White White 20.2 57.2
3 36253 18 years old or older Male 11th Hispanic/Lati… Hisp… NA NA
4 1095530 15 years old Male 10th Black or Afri… Blac… 28.0 85.7
5 1303997 14 years old Male 9th All other rac… Mult… 24.5 66.7
6 926649 16 years old Male 11th All other rac… Asian 20.5 70.3filter_at()
select columns to which the change should happen via the
vars()argument.Use
all_vars()if all columns need to return TRUE, orany_vars()in case just one variable needs to return TRUE.
# A tibble: 3 × 8
record age sex grade race4 race7 bmi stweight
<dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl>
1 1099468 17 years old Male 9th Black or African Ameri… Blac… 52.0 174.
2 770391 16 years old Female 11th All other races Mult… 52.4 161.
3 501097 15 years old Male 12th All other races Mult… 50.3 116.Adding or Modifying columns using mutate()
The dplyr library has the following functions that can be used to add additional variables to a data frame.
mutate()– adds new variables while retaining old variables to a data frame.transmute()– adds new variables and removes old ones from a data frame.mutate_all()– changes every variable in a data frame simultaneously.mutate_at()– changes certain variables by name.mutate_if()– alterations all variables that satisfy a specific criterionThe
mutate()basic syntax is as follows.
-
data: the fresh data frame where the fresh variables will be placed -
new_variable: the name of the new variable -
existing_variable: the current data frame variable that you want to modify in order to generate a new variable
- Set the new column called
height_m
Code
# A tibble: 20,000 × 3
record bmi stweight
<dbl> <dbl> <dbl>
1 931897 17.2 54.4
2 333862 20.2 57.2
3 36253 NA NA
4 1095530 28.0 85.7
5 1303997 24.5 66.7
6 261619 NA NA
7 926649 20.5 70.3
8 1309082 19.3 59.0
9 506337 33.1 123.
10 180494 NA NA
# ℹ 19,990 more rowstransmute()
A data frame’s variables are added and removed via the transmute() method.
The code that follows demonstrates how to eliminate all of the existing variables and add two new variables to a dataset.
mutate_all()
The
mutate_all()function changes every variable in a data frame at once, enabling you to use thefuns()function to apply a certain function to every variable.The use of
mutate_all()to divide each column in a data frame by ten is demonstrated in the code below.divide 10 from each of the data frame’s variables.
# A tibble: 20,000 × 4
record bmi stweight height_m
<dbl> <dbl> <dbl> <dbl>
1 93190. 1.72 5.44 0.178
2 33386. 2.02 5.72 0.168
3 3625. NA NA NA
4 109553 2.80 8.57 0.175
5 130400. 2.45 6.67 0.165
6 26162. NA NA NA
7 92665. 2.05 7.03 0.185
8 130908. 1.93 5.90 0.175
9 50634. 3.31 12.3 0.193
10 18049. NA NA NA
# ℹ 19,990 more rows- Remember that you can add more variables to the data frame by supplying a new name to be prefixed to the existing variable name.
# A tibble: 20,000 × 4
record_mod bmi_mod stweight_mod height_m_mod
<dbl> <dbl> <dbl> <dbl>
1 93190. 1.72 5.44 0.178
2 33386. 2.02 5.72 0.168
3 3625. NA NA NA
4 109553 2.80 8.57 0.175
5 130400. 2.45 6.67 0.165
6 26162. NA NA NA
7 92665. 2.05 7.03 0.185
8 130908. 1.93 5.90 0.175
9 50634. 3.31 12.3 0.193
10 18049. NA NA NA
# ℹ 19,990 more rowsmutate_at()
Using names, the
mutate at()function changes particular variables.The use of
mutate_at()to divide two particular variables by 10 is demonstrated in the code below:
# A tibble: 20,000 × 6
record bmi stweight height_m height_m_mod stweight_mod
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 931897 17.2 54.4 1.78 0.178 5.44
2 333862 20.2 57.2 1.68 0.168 5.72
3 36253 NA NA NA NA NA
4 1095530 28.0 85.7 1.75 0.175 8.57
5 1303997 24.5 66.7 1.65 0.165 6.67
6 261619 NA NA NA NA NA
7 926649 20.5 70.3 1.85 0.185 7.03
8 1309082 19.3 59.0 1.75 0.175 5.90
9 506337 33.1 123. 1.93 0.193 12.3
10 180494 NA NA NA NA NA
# ℹ 19,990 more rowsmutate_if()
All variables that match a specific condition are modified by the
mutate_if()function.The
mutate_if()function can be used to change any variables of type factor to type character, as shown in the code below.every character variable can be converted to a factor variable.
The
mutate_if()method can be used to round any numeric variables to the nearest whole number using the following example code.any numeric variables should be rounded to the nearest decimal place.
# A tibble: 20,000 × 4
age bmi stweight height_m
<chr> <dbl> <dbl> <dbl>
1 15 years old 17.2 54.4 1.8
2 17 years old 20.2 57.1 1.7
3 18 years old or older NA NA NA
4 15 years old 28 85.7 1.7
5 14 years old 24.5 66.7 1.6
6 17 years old NA NA NA
7 16 years old 20.5 70.3 1.8
8 17 years old 19.3 59 1.8
9 18 years old or older 33.1 123. 1.9
10 14 years old NA NA NA
# ℹ 19,990 more rowsUse across() inside mutate() function
# A tibble: 4 × 9
record age sex grade race4 race7 bmi stweight height_m
<dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl>
1 931897 15 years old Female 10th White White 17 54 1.78
2 333862 17 years old Female 12th White White 20 57 1.68
3 36253 18 years old or older Male 11th Hisp… Hisp… NA NA NA
4 1095530 15 years old Male 10th Blac… Blac… 28 86 1.75# A tibble: 4 × 9
record age sex grade race4 race7 bmi stweight height_m
<dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl>
1 931897 15 years old Female 10th White White 17 54 1.78
2 333862 17 years old Female 12th White White 20 57 1.68
3 36253 18 years old or older Male 11th Hisp… Hisp… NA NA NA
4 1095530 15 years old Male 10th Blac… Blac… 28 86 1.75Use across() inside mutate() function
# A tibble: 4 × 9
record age sex grade race4 race7 bmi stweight height_m
<dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl>
1 931897 15 years old Female 10th White White 17 54 2
2 333862 17 years old Female 12th White White 20 57 2
3 36253 18 years old or older Male 11th Hisp… Hisp… NA NA NA
4 1095530 15 years old Male 10th Blac… Blac… 28 86 2# A tibble: 4 × 9
record age sex grade race4 race7 bmi stweight height_m
<dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl>
1 931897 15 years old Female 10th White White 17 54 2
2 333862 17 years old Female 12th White White 20 57 2
3 36253 18 years old or older Male 11th Hisp… Hisp… NA NA NA
4 1095530 15 years old Male 10th Blac… Blac… 28 86 2Sort rows with arrange
Re-order rows by a particular column, by default in ascending order
Use desc() for descending order.
arrange(data, variable1, desc(variable2), ...)
Example: 1. Let us use the following code to create a scrambled version of the airquality dataset
Sort rows with arrange()
Example: 2. Now let us arrange the data frame back into chronological order, sorting by Month then Day
Ozone Solar.R Wind Temp Month Day
1 41 190 7.4 67 5 1
2 36 118 8.0 72 5 2
3 12 149 12.6 74 5 3
4 18 313 11.5 62 5 4
5 NA NA 14.3 56 5 5
6 28 NA 14.9 66 5 6.comment[Try : arrange(air_mess, Day, Month) and see the difference.]
group_by() and summarise()
The
dplyrverbs become especially powerful when they are are combined using the pipe operator|>.The following
dplyrfunctions allow us to split our data frame into groups on which we can perform operations individuallygroup_by(): group data frame by a factor for downstream operations (usually summarise)summarise(): summarise values in a data frame or in groups within the data frame with aggregation functions (e.g.min(),max(),mean(), etc…)
dplyr - Split-Apply-Combine
The group_by function is key to the Split-Apply-Combine strategy
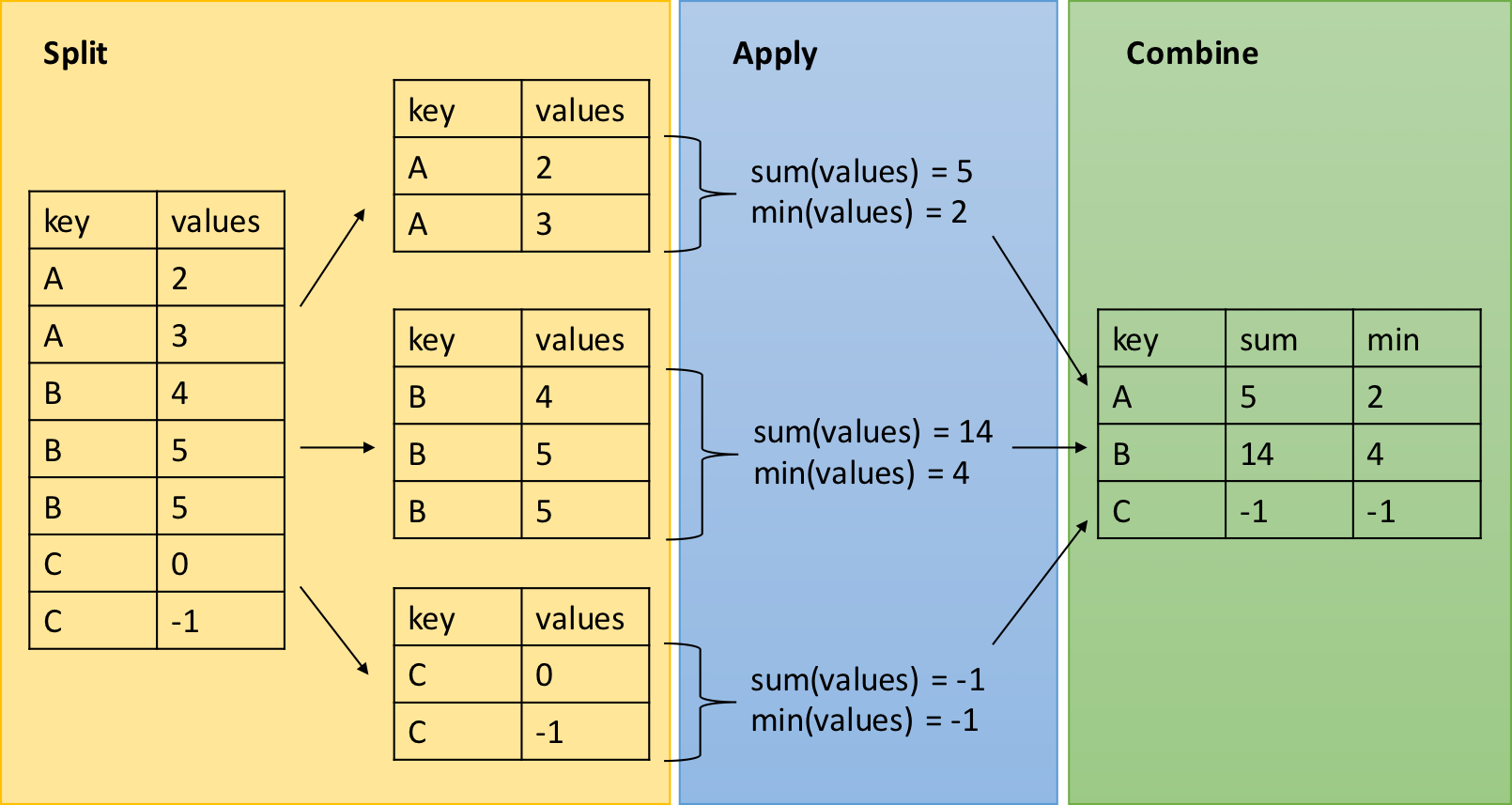
The summarize() function
- The
summarize()function is used in the R program to summarize the data frame into just one value or vector. - This summarization is done through grouping observations by using categorical values at first, using the
group_by()function. - The
summarize()function offers the summary that is based on the action done on grouped or ungrouped data.
dplyr::summarize() Function
- To calculate the mean bmi in base R vs with
summarize()::
Multiple Summary Statistics
You can calculate multiple statistics in one summarize():
# A tibble: 1 × 2
mean_age median_bmi
<dbl> <dbl>
1 23.5 22.3Grouped summaries with dplyr::group_by()
group_by() groups data by one or more variables.
- Example 1: Mean Age by Sex
Result: Different mean ages for each group (e.g., male and female).
- Example 2: Maximum and Minimum Weights
Calculate the max and min weights for each sex:
Code
# A tibble: 2 × 3
sex max_weight min_weight
<chr> <dbl> <dbl>
1 Female 181. 27.7
2 Male 181. 35.4Why summarize() Matters
- The combination of
group_by()andsummarize()allows highly informative grouped summaries of datasets with minimal code.- Producing such summaries is an essential data analysis skill.
Grouping by Multiple Variables (Nested Grouping)
- To group by more than one variable, list both in
group_by():
Code
# A tibble: 4 × 3
# Groups: sex [1]
sex grade mean_bmi
<chr> <chr> <dbl>
1 Female 10th 23.0
2 Female 11th 23.4
3 Female 12th 23.9
4 Female 9th 22.8- You can swap the column order in
group_by():
Ungrouping Data
- After
group_by()andsummarize(), the resulting data frame may still be grouped. - To avoid unintended behaviors, use
ungroup():
Why is ungroup() Needed?
- Grouped data frames behave uniquely with other
dplyrfunctions likeselect(),filter(), ormutate():
Code
# A tibble: 4 × 2
# Groups: sex [1]
sex mean_bmi
<chr> <dbl>
1 Female 23.0
2 Female 23.4
3 Female 23.9
4 Female 22.8- By ungrouping, we get the expected output:
Counting Rows
- Use
n()insidesummarize()to count rows:
Counting Rows with Conditions
- To count rows that meet specific conditions, wrap the condition in
sum():
Code
# A tibble: 8 × 2
race7 count_above50
<chr> <int>
1 Am Indian / Alaska Native 1
2 Asian 1
3 Black or African American 7
4 Hispanic/Latino 2
5 Multiple - Non-Hispanic 3
6 Native Hawaiian/other PI 1
7 White 2
8 <NA> 0- For binary variables,
TRUEequals 1, andFALSEequals 0, makingsum()work seamlessly.
Counting Missing Values
To count NAs:
# A tibble: 3 × 2
sex unknown_bmi
<chr> <int>
1 Female 2970
2 Male 3257
3 <NA> 231To count known (non-missing) values:
Using dplyr::count()
-
count()simplifies counting observations by group:
- You can count by multiple variables:
# A tibble: 15 × 3
sex grade n
<chr> <chr> <int>
1 Female 10th 2332
2 Female 11th 2365
3 Female 12th 2277
4 Female 9th 2492
5 Female <NA> 126
6 Male 10th 2539
7 Male 11th 2496
8 Male 12th 2263
9 Male 9th 2684
10 Male <NA> 195
11 <NA> 10th 36
12 <NA> 11th 30
13 <NA> 12th 37
14 <NA> 9th 43
15 <NA> <NA> 85
count() vs. summarize()
The count() function is limited to row counts, while summarize() can produce multiple summary statistics:
Code
# A tibble: 6 × 4
# Groups: sex [2]
sex grade count median_bmi
<chr> <chr> <int> <dbl>
1 Female 10th 2332 NA
2 Female 11th 2365 NA
3 Female 12th 2277 NA
4 Female 9th 2492 NA
5 Female <NA> 126 NA
6 Male 10th 2539 NASummarize grouped data
The operations that can be performed on grouped data are
average,factor,count,mean, etc.Example: we are interested in the mean temperature and standard deviation within each month of the airquality dataset
Summarize ungrouped data
- We can also summarize ungrouped data. This can be done by using three functions.
summarize_all()summarize_at()summazrize_if()
1. summarize_all()
- This function summarizes all the columns of data based on the action which is to be performed.
summarize_all(action)
- example The code
airquality |> summarize_all(mean)will show the mean of all columns.
2. summarize_at()
It performs the action on the specific column and generates the summary based on that action.
summarize_at(vector_of_columns, action)
vector_of_columns: The list of column names or character vector of column names.
3. summarize_if()
- In this function, we specify a condition and the summary will be generated if the condition is satisfied.
- In the code snippet below, we use the
predicatefunctionis.numericandmeanas an action.